Astex Overlay Pages Help
Contents
Overview
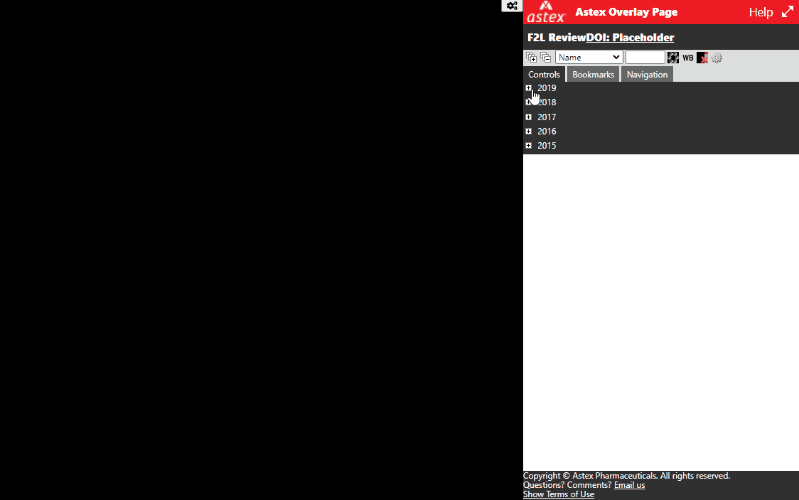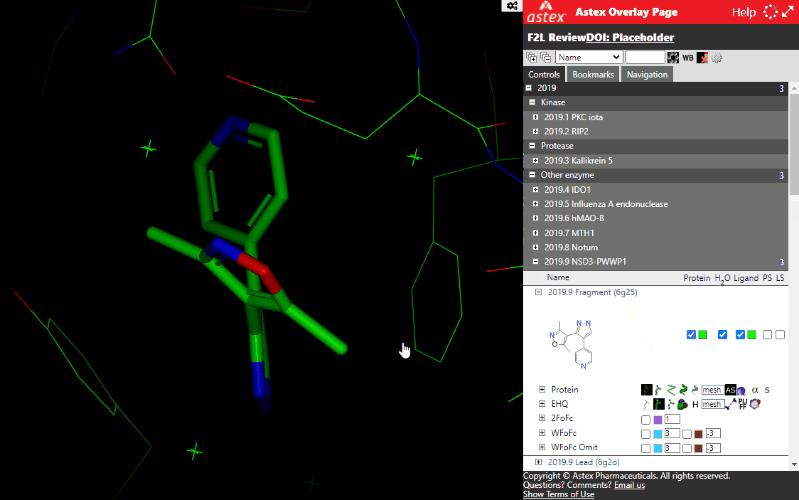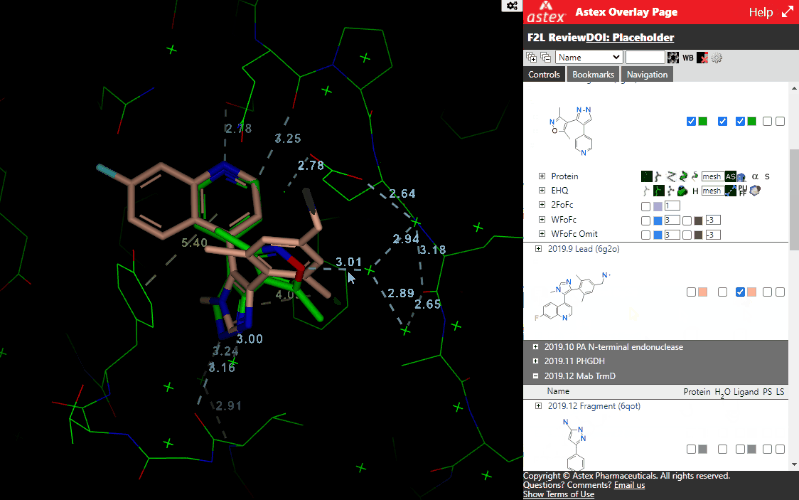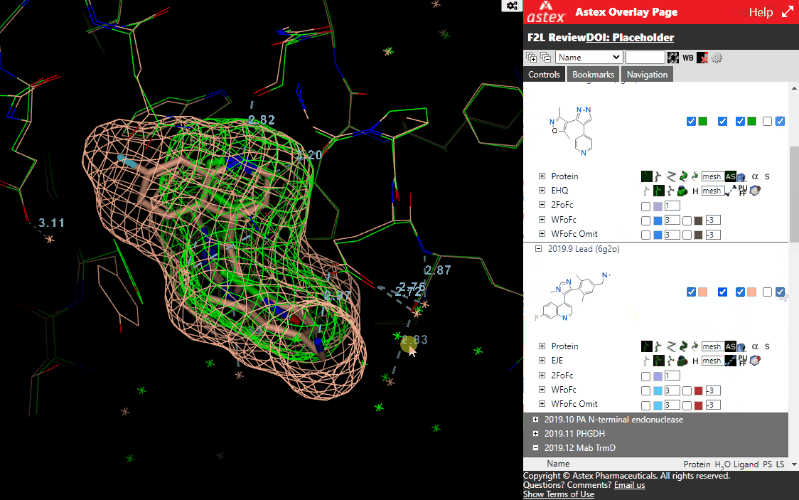
The overlay pages provide a curated view of a series of protein-ligand structures. The structures can be explored and displayed through the hierarchical menus in the right-hand panel.
Structures have some basic top-level controls: checkboxes and colour pickers to control the protein, ligand, waters and simple molecular surfaces.
Expanding a structure displays further controls for different display styles and controls to turn on electron density maps (where available). The maps are often clipped to the immediate vicinity of the ligand to minimize file sizes
UI Controls
Some of the key controls are annotated in the screenshot below:
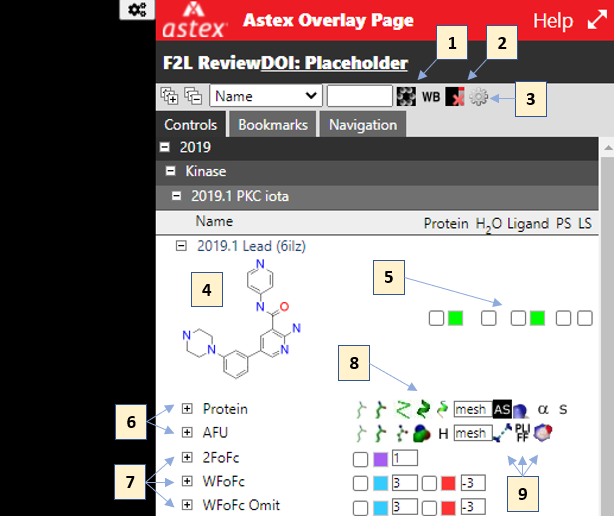
These are:
- Toggle 2D ligand structures
- Clear the viewer (hide everything)
- Open settings menu (includes toggle for dark/light background)
- Clicking on the 2D depiction of a ligand will center the viewer on the ligand
- Top-level controls for a complex, the checkboxes will turn on protein, waters, ligand, protein surface and ligand surface respectively. Clicking on the color swatches opens a color picker
- Expanding a complex reveals rows for each component (protein, ligand etc.). Clicking on the name will center the viewer on that component
- Electron density maps (where available for a structure) are accessed here
- This region has toggles for common representation types, as well as surface settings and a toggle for showing symmetry atoms
- These three controls toggle:
- Noncovalent interactions
- Coloring of protein/ligand/water by PLIFF score (see: Verdonk, ML et al.)
- Ligand AI electrostatic surface calculated using ESP_DNN (see: Rathi, PC et al.)
Bookmarks
Bookmarked views, e.g. for publication figures or presentation are available in the bookmark tab:
Mouse Controls
- Rotate - Left button hold and move
- Zoom - Shift+Left button hold and move, OR Right button hold and drag (up/down)
- Translate - Ctrl+Left button hold and move
- Adjust clipping planes - Scroll mousewheel (OR "-" and "+" keys)
- Pick - Left click on an atom (see measurements below)
- Center -Middle click on an atom or bond
Keyboard Shortcuts
The following keyboard shortcuts are available when the NGL Viewer has focus (i.e. after you click on the viewer area).
General
- (c)enter - recenter on the last picked atom
- (r)eset - zooms to view all loaded structures
- Sp(i)n - toggle spin mode
- Roc(k) - toggle rock mode
- (_-_) - decrease depth-of-field (move clipping planes together) OR mouse scrollwheel up
- (_+_) - increase depth-of-field (move clipping planes apart) OR mouse scrollwheel down
Measurements
Pick to select atoms, then:
- (d)istance - operates on last two picked atoms
- (a)ngle - operates on last three picked atoms
- (t)orsion - operates on last four picked atoms
(Shift-d/a/t) clears distances, angles, torsions respectively
NGL Selection Language
The NGL viewer has a concept of "components" (a protein, ligand, map etc) and representations (lines, ball+stick, surface etc). Representations may be restricted to specific atoms using NGL's selection language - a brief description is available in the NGL documentation.
In the overlay pages, expanding a protein or ligand shows all of the current representations and provides an option to add custom representations. Expanding a representation reveals the full set of parameters that can be modified including the selection.
Contact
If you have any comments or feedback on the overlay pages, please send us an email